ABC - Department of Genomics - Molecular Pant Pathology Group
Molecular Pant Pathology Group
Our group, established in 2013 aims to adapt and develop new, sensitive molecular biology methods based virus diagnostics. We not provide service, but work on basic and applied research, which can establish new diagnostic methods, help us to better know plant viruses and phytoplasmas which knowledge can be used in plant protection strategies. Small RNA HTS is in our focus and we think that virus detection, based on this method will revolutionized virus diagnostics in the next years.
Research interest
Investigation of the effect of virus infection and development of new, high throughput diagnostics methods with increased sensitivity
New, sensitive virus diagnostics based on metagenomic approach.
Virology survey of woody plants: grapevine and fruit trees, description of viruses present in Hungary, investigation of tree decline
Monitoring of the efficiency of the methods for virus release
Optimization and testing of simple, fast, high-throughput virus diagnostic methods.
The aim of work is to develop new, molecular biology techniques (RT-PCR, hybridization methods, next generation sequencing) based detection assays in order to detect viral or phytoplasma infection with high sensitivity and efficiency in grapevine and in fruit trees.
Using these techniques presence of the pathogen can be detected in early, even in latent form and infection what can be the starting point of epidemics on young cultivars can be identified and prevented. We are also interested in molecular mechanism lying behind symptom development in virus-infected plants.
We participate in the COST DIVAS action. http://www.cost.eu/COST_Actions/fa/FA1407
Research projects:
Investigation of plant pathogens causing decline in fruit tree plantations by molecular biology methods(NKFIH K 127951 2018-2022)
Molecular characterization of function, importance and evolution of new emerging grapevine viruses by high throughput sequencing and molecular biology based methods NKFIH K 119783 2016-2020)
Development of virus diagnostics for grapevine and fruit trees and diagnostic monitoring of the production of virus free propagation material. (Cooperation with RIVO and FRI of NARIC)
Molecular mechanism behind disease symptom development in virus infected plants NKFIH K-108718 (2013-2018)
Adaptation of new genomics and biotechnology based techniques in the production of healthy propagation material, plant protection and trait development of grapevine and fruits. KTIA –AIK_12-1-2013-0001 (2013-2015)
Investigation of plant pathogens causing decline in fruit tree plantations by molecular biology methods(NKFIH K 127951 2018-2022)
Apricot orchard of Hungary shows drastic decline in the last few years which were often connected with the widespread presence of phytoplasma (Ca p prunorum). Beside this pathogen presence of new viruses were described by our group causing problems not only on apricot, but also in apple orchards. Metagenomics approaches based next generation sequencing techniques can reveal the presence of all pathogens present in the plant showing symptoms. Presence of bacteria and fungi can disclose by direct genome sequencing, while RNA seq and small RNA NGS can be used to describe viral pathogens in the investigated sample. Further investigation of the complex infections can answer the question which pathogen in which combination cause symptoms leading to severe loss in crop yields. With these new sensitive techniques we can survey not only our orchards but collections of varieties. Our results will also give information about the variants or strains present in our country. This detailed information would help us not only to plan plant protection strategies, but find new avenues to prevent their spread and minimise their damage.
Molecular characterization of function, importance and evolution of new emerging grapevine viruses by high throughput sequencing and molecular biology based methods NKFIH K 119783 2016-2020)
There are more than 60 viral pathogens (virus or viroid) infecting grapevine. Viral diseases cannot be controlled by traditional plant protection methods, their infection can be prevented only by the usage of virus free propagating material. Discovery of RNA interference and fast evolution in sequencing techniques opens new possibilities for diagnostics methods. Deep sequencing of small RNA libraries offered unique opportunity to reveal the presence of all viral pathogens in the sample. During our previous project, using this cutting edge technique, we identified two new viruses in Hungarian grapevine plantations not described from Hungary before and not characterized molecularly.
The aim of the current project is the molecular characterization of these two viruses to support improvement of their diagnostics and new approaches for the production of clean propagating material to prevent their spread. Their phylogenetically relation to their known relatives will revealed. Viral proteins encoded by them will be investigated in order to identify their possible viral silencing suppressor. In situ hybridization of tissues at different developmental stages of grapevine will reveal spatial and temporal pattern of their distribution and symptom development in infected plants. New diagnostics methods, based on RNA detection, will be developed and used to survey their presence in Hungarian rootstock plantations. Their movement will be monitored in grafts using virus infected rootstock. Regular and newly developed protocols for virus free propagation material production will be optimized for their enhanced efficiency against these new viruses.
Development of virus diagnostics for grapevine and fruit trees and diagnostic monitoring of the production of virus free propagation material. (Cooperation with RIVO and FRI of NARIC)
Aim of our project is to reveal virus infection status of Hungarian fruit trees and to determine what viruses are important and have serious problems in Hungary. Based on these results we will be able to adopt and develop new molecular biology based approaches what are fast and sensitive to detect virus infection. The optimized protocol will allow us to monitor different steps of the virus free propagation material production process, to make it more effective.
Molecular mechanism behind disease symptom development in virus infected plants NKFIH K-108718 (2013-2018)
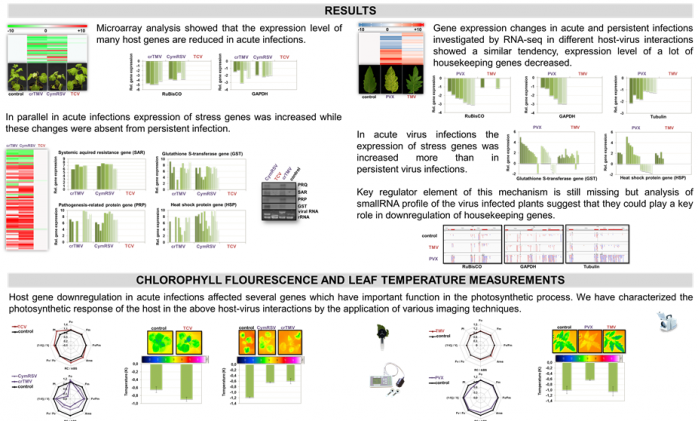
In compatible plant-virus infection the virus can invade the whole plant forming the disease symptoms specific for the particular plant-virus interaction generating severe economic loss. Successful viral infection and development of disease symptoms are mainly determined by the efficiency of virus spread in the plant and the magnitude of virus induced changes on the host metabolism. Movement and spreading of the virus is efficiently controlled by RNA silencing based host defence, while severity of gene expression changes depends on the type of the plant-virus connection. Previously we revealed that most of the viruses can control one of the key molecules, the ARGONAUTE 1 (AGO1) protein, of the gene silencing machinery via the regulation of a specific microRNA. AGO1 plays a central role in the miRNA mediated essential developmental regulation and can be an important component of the plant defence reaction against viruses. In our current project, we investigate the complex regulation pattern of AGO1, its role in symptom development and in defence reactions against different viruses. Furthermore, we analyse the additional role of other, not AGO1 connected, gene-expression changes (identified by microarray analyses in our previous OTKA project) in the determination of the severity of disease symptoms. As a result, we will gain a comprehensive knowledge of disease symptom development during virus infection in Arabidopsis model plant and in Solanaceae species. Our knowledge might help us to find alternative possibilities to protect plants against different viruses or alleviate the economic loss of virus infections.
Adaptation of new genomics and biotechnology based techniques in the production of healthy propagation material, plant protection and trait development of grapevine and fruits. KTIA –AIK_12-1-2013-0001 (2013-2015)
Usage of genomic information can be crucial in plant breeding and in cultivation techniques. Based on this knowledge we can develop traits with increased pathogen resistance, higher and better yields and with increased adaptation potential to different biotic and abiotic stresses. Traditional breeding methods can be dramatically accelerated by the help of up-to date genomic information and investigation.
Our project opened the possibilities of state of the art next generation genom, transcriptome and small RNA sequencing methods in parallel with newest bioinformatics pipelines to the Hungarian agriculture to support both basic research, breeding programs and production of pathogen free propagation material.
In the agronext project, we monitored pathogen infection of stock cultivations of grapevine and fruits from different part of Hungary focusing on the presence of viruses and viroids by small RNA sequencing at Illumina platform.
This work was a collaboration of Diagnostic Group, Virology Group of NARIC ABC and Department of Viticulture of the Institute of Viticulture and Oenology at Corvinus University of Budapest and coordinated by Burgyán József.
DIAGNOSTiC GROUP 2019
DIAGNOSTiC GROUP 2018
DIAGNOSTiC GROUP 2017
DIAGNOSTiC GROUP 2015-2016
DIAGNOSTiC GROUP 2013-2014
Staff
Publications
Publications
2018
in periodicals:
Nikoletta Czotter, Janos Molnar, Emese Szabó, Emese Demian, Levente Kontra, Ivett Baksa, Gyorgy Szittya, Laszlo Kocsis, Tamas Deak, Gyorgy Bisztray, Gabor E. Tusnady, Jozsef Burgyan and Eva Varallyay (2018) NGS of virus-derived small RNAs as a diagnostic method used to determine viromes of Hungarian vineyards. Frontiers in Microbiology 9, 122 (2018) doi.org/10.3389/fmicb.2018.00122
Czotter, N., Molnár, J., Pesti, R., Demián, E., Baráth, D., Varga, T., Várallyay, É. (2018) Use of siRNAs for Diagnosis of Viruses Associated to Woody Plants in Nurseries and Stock Collections. In: Viral Metagenomics: Methods and Protocols. (Pantaleo, V. and Chiumenti, M., eds.). New York, NY: Springer New York, pp. 115-130. doi.org/10.1007/978-1-4939-7683-6_9
Baráth, D.; Jaksa-Czotter, N.; Molnár, J.; Varga, T.; Balássy, J.; Szabó, L.K.; Kirilla, Z.; Tusnády, G.E.; Preininger, É.; Várallyay, É. Small RNA NGS Revealed the Presence of Cherry Virus A and Little Cherry Virus 1 on Apricots in Hungary. Viruses 2018, 10, 318. doi.org/10.3390/v10060318
Massart, S., Chiumenti, M., De Jonghe, K., Glover, R., Haegeman, A., Koloniuk, I., Komínek, P., Kreuze, J., Kutnjak, D., Lotos, L., Maclot, F., Maliogka, V.I., Maree, H., Olivier, T., Olmos, A., Pooggin, M., Reynard, J.-S.S., Ruiz-García, A.B., Safarova, D., Schneeberger, P.H.H., Sela, N., Turco, S., Vainio, E.J., Varallyai, E., Verdin, E., Westenberg, M., Brostaux, Y. and Candresse, T. (2018) Virus detection by high-throughput sequencing of small RNAs: large scale performance testing of sequence analysis strategies. Phytopathology. 2018 Aug 2. doi: 10.1094/PHYTO-02-18-0067-R.
at conferences:
Emese Demián, Nikoletta Czotter and Éva Várallyay (2018) Detection of Grapevine Pinot gris Virus in different non-Vitis hosts in Hungary. 19th Conference of the ICVG, Santiago of Chile, 9-12 of April, 2018.
Levente Kontra, Emese Demián, Nikoletta Czotter, János Lázár, Lehoczky János and Éva Várallyay (2018) GLPV – an unknown, but familiar grapevine virus. 19th Conference of the ICVG, Santiago of Chile, 9-12 of April, 2018.
Daniel Barath, David Czako, Rozalia Zsovakne-Hangyal, Tunde Varga, Aliz Furton, Mohammad Omran, Klara Nyerges and Eva Varallyay (2018) Comparison of biotest and smallRNA NGS as a virus diagnostics method on fruit trees. Final Meeting of COST-DIVAS Action: HTS Technologies for the study and diagnostic of plant viruses, Liège (Belgium) 26th to 30th November 2018.
Dániel Baráth, Nikoletta Czotter, Alexandra Bükki, Luca Szabó, Zoltán Kirilla, Éva Preininger, Éva Várallyay (2018) First report of Peach latent mosaic viroid in Hungary. Advances in Plant Virology, Birmingham, Birmingham, UK 1April 11-13, 2018.
Demián Emese, Nikoletta Czotter, Éva Várallyay (2018) Investigation of Grapevine Pinot gris Virus presence in non-Vitis hosts in Hungary. Power of Viruses. International Conference, Poreč, Croatia, May 16-18, 2018.
Dániel Baráth, Alexandra Bükki, Dávid Czakó, Nikoletta Czotter, Luca Krisztina Szabó, Zoltán Kirilla, Éva Preininger and Éva Várallyay (2018) Detection of CVA, LChV1 and PLMVd in Prunus sps in Hungarian stock collections. International Conference, Poreč, Croatia, May 16-18, 2018.
2017
at conferences:
Nikoletta Czotter, Tünde Varga, Dániel Baráth, János Molnár, Júlia Balássy, Tamás Deák, Gábor E. Tusnády, László Kocsis, Éva Preininger, József Burgyán and Éva Várallyay (2017) NGS studies on fruit tree screenhouses and stock collections in Hungary 24th International Conference on Virus and Other Graft Transmissible Diseases of Fruit Crops, Thessalloniki, 2017 junius 5-9.
Nikoletta Czotter, Emese Demián, János Molnár , Tamás Deák, Gábor E. Tusnády, László Kocsis, József Burgyán, Éva Várallyay (2017) Virus diagnosics of Hungarian grapevine plantations by next generation sequencing of small RNAs 3rd Hungarian Molecular Life Sciences Conference Eger, 2017.március 31-április 2.
Réka Pesti, Levente Kontra, Kenny Paul, János Molnár, Gábor E. Tusnády, Imre Vass, Zoltán Havelda, Éva Várallyay (2017) Characterization of gene expression and physiological changes in different host-virus interactions 3rd Hungarian Molecular Life Sciences Conference Eger, 2017.március 31-április 2.
Dániel Baráth, Nikoletta Czotter, Júlia Balássy, Tünde Varga, Luca Szabó, Zoltán Kirilla, János Molnár, Gábor E. Tusnády, Éva Preininger, Éva Várallyay (2017) Undescribed viroid detection by metagenomic approach in Hungary 3rd Hungarian Molecular Life Sciences Conference Eger, 2017.március 31-április 2.
Emese Demián, Nikoletta Czotter, Kamilla Czakó, János Molnár, Gábor E. Tusnády, Éva Várallyay (2017) Detection and molecular characterization of Hungarian strain of Grapevine Pinot gris Virus 3rd Hungarian Molecular Life Sciences Conference Eger, 2017.március 31-április 2.
Réka Pesti, Oláh Enikő, Ferenc Kagan, Zoltán Havelda, Várallyay Éva (2017) Sequence requirement of viral suppressor mediated miR168 induction in plants 3rd Hungarian Molecular Life Sciences Conference Eger, 2017.március 31-április 2.
Sebastien Massart, Ian Adams, Kris De Jonghe, Annalisa Giampetruzzi, Igor Koloniuk, Petr Kominek, Jan Kreuze, Denis Kutnjak, Leonidas Lotos, Hans J. Maree, Thibaut Olivier, Mikhail Pooggin, Ana B. Ruiz-García, Dana Safarova, P. H. H. Schneeberger, Noa Sela, Eva Varallyay, Eeva Vainio, Eric Verdin, Marcel Westenberg, Yves Brostaux and Thierry Candresse (2017) High Throughput Sequencing of siRNAs and virus diagnostic: does sequence analysis strategies really matter? Results of an international proficiency testing 3rd Hungarian Molecular Life Sciences Conference Eger, 2017.március 31-április 2., 24th International Conference on Virus and Other Graft Transmissible Diseases of Fruit Crops, Thessalloniki, 2017 junius 5-9.
2016
in periodicals:
Enikő Oláh, Réka Pesti, Dénes Taller, Zoltán Havelda and Éva Várallyay (2016) Non-targeted effects of VIGS vectors on host endogenous gene expression, Archives of Virology 161:2387–2393 (doi:10.1007/s00705-016-2921-9) IF:2,39
Endre Barta, Zsófia Bánfalvi, Zoltán Havelda, László Hiripi, Zoltán Jeney, János Kiss, Balázs Kolics, Ferenc Marincs, Dániel Silhavy, Éva Várallyay (2016) Agricultural genomics: an overview of next generation sequencing projects at the NARIC-Agricultural Biotechnology Institute, Hungarian Agricultural Research 25:10-21
at conferences:
Czotter N, Molnár J, Tusnády E. G, Kocsis L, Burgyán J, Várallyay É. (2016) Validation of smallRNA NGS based virus diagnostics by RT-PCR, International Advances in Plant Virology, Conference of AAB in conjunction with Cost Action FA1407, Greenwich, 2016 Sept 6-9
Czotter N, Molnár J, Deák T, Tusnády E. G, Kocsis L, Burgyán J, Várallyay É. (2016) Virus diagnosis in grapevine plantations with next generation sequencing metagenomic approach, International Advances in Plant Virology, Conference of AAB in conjunction with Cost Action FA1407, Greenwich, 2016 Sept 6-9,
Pesti Réka, Molnár János, Kenny Paul, Vass Imre, Tusnády E.Gábor, Havelda Zoltán, Várallyay Éva (2016) Characterization of gene expression and physiological changes in different host-virus interactions, International Advances in Plant Virology, Conference of AAB in conjunction with Cost Action FA1407, Greenwich, 2016 Sept 6-9, Poster award
Sebastien Massart, Antonio Olmos, Ian Adams, Kris De Jonghe, Annalisa Giampetruzzi, Igor Koloniuk, Petr Kominek, Jan Kreuze, Denis Kutnjak, Varvara Maliogka, Hano Maree, Thibaut Olivier, Mikhail Pooggin, Dana Safarova, Pierre Schneeberger, Noa Sela, Candresse Thierry, Eva Varallyay, Eeva Vainio, Eric Verdin, Marcel Westenberg and Yves Brostaux (2016) High Throughput Sequencing and virus diagnostic: does bioinformatics really matter? Results of an international bioinformatic proficiency testing, International Advances in Plant Virology, Conference of AAB in conjunction with Cost Action FA1407, Greenwich, 2016 Sept 6-9
2015
in periodicals:
Czotter, E. Szabó, J. Molnár, L. Kocsis, J., T. Deák, Gy. Bisztray, G. E. Tusnády, J. Burgyán, É. Várallyay. (2015) First description of Grapevine Syrah virus-1 in vineyards of Hungary, Journal of Plant Pathology 97 (S), S74
Czotter, N., Manduláné Farkas, E., Lózsa, R., Ember, I.,Szûcsné Varga, G., Várallyay, É. ,Szegedi, E(2015). Primers designed for the detection of grapevine pathogens spreading with propagating material by quantitative real-time PCR . International Journal of Horticultural Science 21 (1–2): 21–30.
at conferences:
Várallyay, É, Oláh, E, Havelda, Z. (2015) Independent parallel functions of p19 plant viral supressor of RNA silencing required for effective supressor activity. Hungarian Molecular Life Sciences 2015, 27-29 March, Eger
Czotter, E. Szabó, J. Molnár, L. Kocsis, J. Lázár, E. Szegedi, T. Deák, Gy. Bisztray, G. E. Tusnády, J. Burgyán, É. Várallyay. (2015) Viromes of Hungarian grapevine plantations by next generation sequencing of small RNAs. 18th Congress of the ICVG, Ankara, 2015 Sept 7-11.
Miklós Makay, Éva Preininger, Éva Várallyay (2015) Importance and solutions for clean stocks: virus diagnostics, in vitro propagation and distribution. Knowledge Exchange and Coordination Workshop on Agricultural Biotechnology, Gödöllő, 2015 okt 27-29.
Pesti, R., Havelda, Z., Várallyay, É (2015) Gene expression changes behind symptom development in virus infected plants. Hungarian Molecular Life Sciences 2015, 27-29 March, Eger
Czotter, N, Szabó E, Nagy T, Deák T, Lázár J, Barta E, Tóth G, Bisztray Gy, Burgyán J, Várallyay É. (2015) NGS aided diagnostics revealed the presence of recently described viruses in Hungarian grapevine plantations. Hungarian Molecular Life Sciences 2015, 27-29 March, Eger
before 2015
in periodicals:
E.Farkas, N. Czotter, R. Lózsa, T. Dula, I. Ember, É. Várallyay, E. Szegedi (2014) Conventional PCR primers for the detection of grapevine pathogens disseminated by propagating material.International Journal of Horticultural Sciences 20(3-4):69-80
Éva Várallyay, Enikő Oláh and Zoltán Havelda (2014): Independent paralleled functions of p19 plant viral suppressor of RNA silencing required for effective suppressor activity. Nucleic Acids Res. 2013 Sep 22. 42: pp. 599-608.
Éva Várallyay and Zoltán Havelda (2013): Unrelated viral suppressors of RNA silencing mediate the control of ARGONAUTE1 level . Molecular Plant Pathology 2013 Aug;14(6):567-75. doi: 10.1111/mpp.12029. Epub 2013 Apr 11.
Várallyay E, Giczey G., Burgyán J. (2012) Virus induced gene silencing of Mlo genes induces powdery mildew resistance in Triticum aestivum. Arch Virol 157:1345–1350
Várallyay E, Válóczi A, Agyi A, Burgyán J and Havelda Z (2010) Plant virus-mediated induction of miR168 is associated with repression of ARGONAUTE1 accumulation. The EMBO Journal , 20;29(20):3507-19.
Varallyay E*, Lichner Z, Safrany J, Havelda Z, Salamon P, Bisztray G, Burgyan J. (2010) Development of a virus induced gene silencing vector from a legumes infecting tobamovirus. Acta Biologica Hungarica, 2010 61(4) 457-469
Havelda Z, Varallyay E, Valoczi A and Burgyan J (2008) Plant virus infection-induced persistent host gene downregulation in systemically infected leaves. Plant Journal, 55:(2) pp. 278-288. (IF:6,75)
Várallyay E., Burgyan J. and Havelda Z. (2008) MicroRNA detection by northern blotting using locked nucleic acid probe. Nature Protocols, 3:(2) 190-196.
Várallyay E., Burgyan J. and Havelda Z. (2007) Detection of microRNAs by northern blot analyses using LNA probes. METHODS: A COMPANION TO METHODS IN ENZYMOLOGY, 43 (2) : 140-45
Valoczi A, Varallyay E, Kauppinen S, Burgyan J, Havelda Z. (2006) Spatio-temporal accumulation of microRNAs is highly coordinated in developing plant tissues. Plant J. 47(1):140-51.
Molnar A, Csorba T, Lakatos L, Varallyay E, Lacomme C, Burgyan J.(2005)
Plant virus-derived small interfering RNAs originate predominantly from highly structured single-stranded viral RNAs. Journal of Virology 79(12):7812-8.