Kutatás
AZ ÁLLATBIOTECHNOLÓGIA FŐOSZTÁLY LEGFRISSEBB PUBLIKÁCIÓI
A GENETIKA FŐOSZTÁLY FŐOSZTÁLY LEGFRISSEBB PUBLIKÁCIÓI
A GENOMIKA FŐOSZTÁLY LEGFRISSEBB PUBLIKÁCIÓI
A NÖVÉNYBIOTECHNOLÓGIA FŐOSZTÁLY LEGFRISSEBB PUBLIKÁCIÓI
AZ ÁLLATBIOTECHNOLÓGIA FŐOSZTÁLY LEGFRISSEBB PUBLIKÁCIÓI
DOI: 10.1016/j.mce.2018.11.002 |
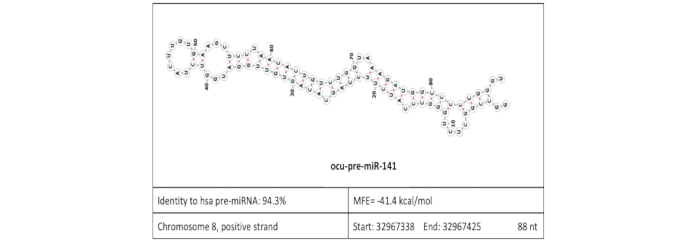
Trophoblastic microRNAs are downregulated in a diabetic pregnancy through an inhibition of Drosha
Pendzialek SM, Knelangen JM, Schindler M, Gürke J, Grybela KJ, Gocza E, Fischer B, Navarrete Santos A
MicroRNAs are promising biological markers for prenatal diagnosis. They regulate placental development and are present in maternal plasma. Maternal metabolic diseases are major risk factors for placental deterioration. Maternal diabetes led to a marked downregulation of Dicer protein in embryoblast cells and Drosha protein in trophoblast cells. MiR-27b, miR-141 and miR-191 were decreased in trophoblast cells and in maternal plasma of diabetic rabbits. As the altered microRNA expression was detectable in maternal plasma, too, the plasma microRNA signature could serve as an early biological marker for the prediction of trophoblast function during a diabetic pregnancy.
DOI: 10.1556/004.2018.046 |
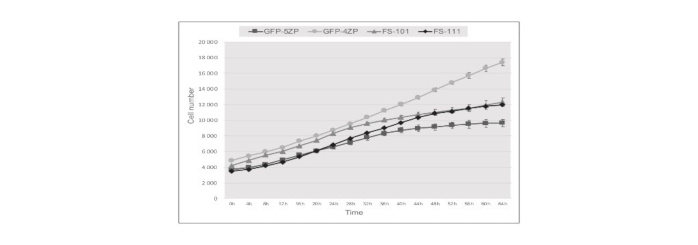
Enhancement of chicken primordial germ cell in vitro maintenance using an automated cell image analyser.
Anand M, Lázár B, Tóth R, Páll E, Patakiné Várkonyi E, Liptói K, Homolya L, Hegyi Z, Hidas A, Gócza E.
We established four PGC lines: two males (GFP-4ZP, FS101) and two females (GFP-5ZP, FS111). We could not detect a significant difference in the marker expression profile, but there was a remarkable difference between the proliferation rate of these PGC lines. We monitored the number of PGCs throughout a three-day period using a high content screening cell imaging and analyzing system (HCS). We compared three different initial cell concentrations in the wells. We could conclude that the difference in proliferation rate could be accounted mainly for the genotypic variation in the established PGC lines, but external factors such as cell concentration and quality of the culture medium also affect the growth rate of PGCs.
DOI: 10.1155/2018/1780679 |
Comparison of the MicroRNA Expression Profiles of Male and Female Avian Primordial Germ Cell Lines
DOI: 10.1556/004.2018.026 |
Glomerulosclerosis in transgenic rabbits with ubiquitous Venus protein expression
Nándor Lipták, Orsolya Ivett Hoffmann, Gabriella Skoda, Elen Gócza, Andrea Kerekes, Zsuzsanna Bősze, László Hiripi
DOI: 10.1371/journal.pone.0187214. |
Secretion of a recombinant protein without a signal peptide by the exocrine glands of transgenic rabbits.
Kerekes A, Hoffmann OI, Iski G, Lipták N, Gócza E, Kues WA, Bősze Z, Hiripi L.
Transgenic rabbits carrying mammary gland specific gene constructs are extensively used for excreting recombinant proteins into the milk. Here, we report refined phenotyping of previously generated Venus transposon-carrying transgenic rabbits with particular emphasis on the secretion of the reporter protein by exocrine glands, such as mammary, salivary, tear and seminal glands. The Sleeping Beauty (SB) transposon transgenic construct contains the Venus fluorophore cDNA, but without a signal peptide for the secretory pathway, driven by the ubiquitous CAGGS (CAG) promoter. Despite the absence of a signal peptide, the fluorophore protein was readily detected in milk, tear, saliva and seminal fluids. The expression pattern was verified by Western blot analysis. Mammary gland epithelial cells of SB-CAG-Venus transgenic lactating does also showed Venus-specific expression by tissue histology and fluorescence microscopy. In summary, the SB-CAG-Venus transgenic rabbits secrete the recombinant protein by different glands. This finding has relevance not only for the understanding of the biological function of exocrine glands, but also for the design of constructs for expression of recombinant proteins in dairy animals.
DOI: 10.1371/journal.pone.0185662. |
Placenta-specific gene manipulation in rabbits.
Skoda G, Hoffmann OI, Gócza E, Bodrogi L, Kerekes A, Bösze Z, Hiripi L.
Lentiviral gene constructs can be efficiently and specifically delivered to trophoblast cell lineages in rodents. In vivo genetic manipulation of trophoblast cell lines enables functional and developmental studies in the placenta. In this report we show that genetic modification can be produced in the extraembryonic tissues of rabbits by lentiviral gene constructs. When 8-16 cell stage embryos were injected with lentiviral particles, strong reporter gene expression resulted in the rabbit placenta. The expression pattern displayed some mosaicism. A strikingly high degree of mosaic GFP expression was detected in some parts of the yolk sac, which is a hypoblast-derived tissue. Whereas expression of the reporter gene construct was detected in placentas and yolk sacs, fetuses never expressed the transgene. As rabbits are an ideal model for functional studies in the placenta, our method would open new possibilities in rabbit biotechnology and placentation studies.
DOI: 10.1007/s11248-016-9994-9 |
|
Monitoring of Venus transgenic cell migration during pregnancy in non-transgenic rabbits |
Kísérleteink során három módszerrel (konfokális mikroszkópia, áramlási citometria, kvantitatív polimeráz láncreakció) vizsgáltuk a sejtvándorlást terhesség alatt és után, Venus transzgénikus és vad típusú nyulakban és utódaikban. Nem találtunk fluoreszcens transzgénikus sejteket a vad típusú nyulakban, vagyis nem volt sejtvándorlás a magzatok és az anyjuk között. Eredményeink arra is rávilágítottak, hogy nincs kimutatható GM kontamináció vad típusú nyulakban és utódaikban transzgénikus nyulakkal történő keresztezés esetén.
Cell transfer between mother and fetus were demonstrated previously in several species which possess haemochorial placenta (e.g. in humans, mice, rats, etc.). Here we report the assessment of fetal and maternal microchimerism in non-transgenic (non-TG) New Zealand white rabbits which were pregnant with transgenic (TG) fetuses and in non-TG newborns of TG does. The TG construct, including the Venus fluorophore cDNA driven by a ubiquitous cytomegalovirus enhancer, chicken ß-actin promoter (CAGGS), was previously integrated into the rabbit genome by Sleeping Beauty transposon system. Three different methods [fluorescence microscopy, flow cytometry and quantitative polymerase chain reaction (QPCR)] were employed to search for TG cells and gene products in blood and other tissues of non-TG rabbits. Venus positive peripheral blood mononuclear cells (PBMCs) were not detected in the blood of non-TG littermates or non-TG does by flow cytometry. Tissue samples (liver, kidney, skeletal and heart muscle) also proved to be Venus negative examined with fluorescence microscopy, while histology sections and PBMCs of TG rabbits showed robust Venus protein expression. In case of genomic DNA (gDNA) sourced from tissue samples of non-TG rabbits, CAGGS promoter-specific fragments could not be amplified by QPCR. Our data showed the lack of detectable cell transfer between TG and non-TG rabbits during gestation.
DOI: Közlésre elfogadva |
| Comparison the microRNA expression profiles of male and female avian primordial germ cell lines. Lazar B, Anand M, Tóth, R, Várkonyi EP, Krisztina Liptói, Gócza E. |
Primordial germ cells (PGCs) are the precursors of adult germ cells and among the embryonic stem-like cells in the bird embryo only they can transmit the genetic information to the next generation. As a first step, we compared 12 newly established chicken PGC lines derived from two different chicken breeds, performing CCK-8 proliferation assay. As microRNAs have been proved to play a key role in the maintenance of pluripotency and the cell cycle regulation of stem cells, we continued with a complex miRNA analysis. We could discover miRNAs expressing differently in PGC lines with high proliferation rate, compared to PGC lines with low proliferation rate. We found that gga-miR-2127 express differently in female and male cell lines. The microarray analysis also revealed high expression level of the gga-miR-302b-3p strand (member of miR-302/367 cluster) in slowly proliferating PGC lines compared to gga-miR-302b-5p strand. We confirmed that the inhibition of miR-302b-5p significantly increases the doubling time of the examined PGC lines.
In conclusion, we found that gga-miR-181-5p, gga-miR-2127 and members of the gga-miR-302/367 cluster have a dominant role in the regulation of avian primordial germ cell proliferation.
DOI: Közlésre elfogadva |
|
Glomerulosclerosis in transgenic rabbits with ubiquitous Venus protein expression Nándor Lipták, Orsolya Ivett Hoffmann, Gabriella Skoda, Elen Gócza, Andrea Kerekes, Zsuzsanna Bősze, László Hiripi |
Kutatásaink során enyhe glomerulosclerosist (enyhe proteinuria, hematuria, glomerulomegália) mutattunk ki Venus transzgénikus nyulakban, míg azonos korú vad típusú egyedekben ezek a tünetek nem jelentkeztek. Korábban csak GFP transzgénikus egerekben detektáltak hasonló tüneteket.
DOI: 10.1556/004.2017.047 |
X- and Y-chromosome-specific variants of the amelogenin gene allow non-invasive sex diagnosis for the detection of pseudohermaphrodite goats.
Fábián R, Kovács A, Stéger V, Frank K, Egerszegi I, Oláh J, Bodó S.
The goat (Capra aegagrus f. hircus) is an important livestock animal for multiple purposes in agricultural production. The Polled Intersex Syndrome (PIS) is responsible for the absence of horns in homozygous and heterozygous goats causing a female-to-male sex-reversal in the homozygous polled genotypic female (XX) goats. We elaborated a simple and efficient non-invasive method to detect the genotypic sex from hair and fecal samples using a pair of primers to amplify the X- and Y-linked alleles of the amelogenin gene. The PCR products were easily distinguishable using agarose gel electrophoresis: at the samples originated from healthy phenotypic females we detected an X-specific single band and at the males double (X- and Y-) bands. The new PCR method is applicable to diagnose the sex of PIS affected animals already at newborn kids, contrary to the phenotypic finds appearing after puberty and may replace the cumbersome chromosome investigations.
DOI: 10.1007/s10499-018-0250-6 |
Immunogene expression in head kidney and spleen of common carp (Cyprinus carpio L.) following thermal stress and challenge with Gram-negative bacterium, Aeromonas hydrophila.
Shahi N, Ardo L, Fazekas G, Gocza E, Kumar S, Revesz N, Sandor ZJ, Molnar Z, Jeney G, Jeney Z.
To evaluate the effect of thermal and microbial stress on the immune response of common carp (Cyprinus carpio L.), relative mRNA expression level of pro-inflammatory cytokines [tumor necrosis factor alpha (TNF-α) and interleukin (IL)-1β] and other genes related to immune or stress response [inducible nitric oxide synthase (iNOS), heat shock protein 70 (Hsp70), superoxide dismutase one (SOD1), and glucocorticoid receptor (GR)] was measured by quantitative PCR (qPCR). In addition, total protein and total immunoglobulin level in blood plasma of experimental common carp was also assayed. All the above parameters were estimated 24 h post-challenge with Gram-negative bacterium, Aeromonas hydrophila. Common carp (54.89 ± 6.90 g) were initially exposed to 20 °C (control group) and 30 °C (thermal stress group) water temperature for 30 days, followed by experimental challenge with 2.29 × 108 colony forming unit/mL (CFU/mL; LD50 dose) of A. hydrophila. Exposure of fish to thermal stress and subsequently challenge with A. hydrophila significantly (P < 0.05) increases the IL-1β mRNA expression in head kidney and spleen of common carp by ~39.94 and ~4.11-fold, respectively. However, TNF-α mRNA expression in spleen decreased ~ 5.63-fold in control fish challenged with A. hydrophila. Thermal stress and challenge with bacterium suppresses the iNOS and GR mRNA expression in spleen of common carp. Moreover, significant (P < 0.05) increase in total protein content of blood plasma (~ 43 mg/g) was evident in fish exposed to thermal stress and challenged with A. hydrophila. In conclusion, our study highlights the importance of elevated temperature stress and microbial infection in differential regulation of expression of several immunogenes in common carp.
A GENETIKA FŐOSZTÁLY FŐOSZTÁLY LEGFRISSEBB PUBLIKÁCIÓI
DOI: 10.3389/fmicb.2019.00457 |
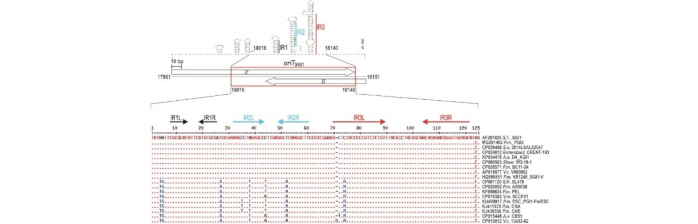
Identification and Characterization of oriT and Two Mobilization Genes Required for Conjugative Transfer of Salmonella Genomic Island 1
János Kiss, Mónika Szabó, Anna Hegyi, Gregory Douard, Karine Praud, István Nagy, Ferenc Olasz, Axel Cloeckaert, Benoît Doublet
The integrative mobilizable elements of SGI1-family considerably contribute to the spread of resistance to critically important antibiotics among enteric bacteria. Even though many aspects of SGI1 mobilization by IncA and IncC plasmids have been explored, the basic transfer elements such as oriT and self-encoded mobilization proteins remain undiscovered. Here we describe the mobilization region of SGI1 that is well conserved throughout the family and carries the oriTSGI1 and two genes, mpsA and mpsB (originally annotated as S020 and S019, respectively) that are essential for the conjugative transfer of SGI1. OriTSGI1, which is located in the vicinity of the two mobilization genes proved to be a 125-bp GC-rich sequence with several important inverted repeat motifs. The mobilization proteins MpsA and MpsB are expressed from a bicistronic mRNA, although MpsB can be produced from its own mRNA as well. The protein structure predictions imply that MpsA belongs to the lambda tyrosine recombinase family, while MpsB resembles the N-terminal core DNA binding domains of these enzymes. The results suggest that MpsA may act as an atypical relaxase, which needs MpsB for SGI1 transfer. Although the helper plasmid-encoded relaxase proved not to be essential for SGI1 transfer, it appeared to be important to achieve the high transfer rate of the island observed with the IncA/IncC-SGI1 system.
DOI: 10.1093/nar/gky279. |
|
The nonstop decay and the RNA silencing systems operate Szádeczky-Kardoss I, Csorba T, Auber A, Schamberger A, Nyikó T, |
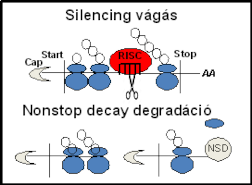
Translation-dependent mRNA quality control systems protect the protein homeostasis of eukaryotic cells by eliminating aberrant transcripts and stimulating the decay of their protein products. Although these systems are intensively studied in animals, little is known about the translation-dependent quality control systems in plants. Here, we characterize the mechanism of nonstop decay (NSD) system in Nicotiana benthamiana model plant. We show that plant NSD efficiently degrades nonstop mRNAs, which can be generated by premature polyadenylation, and stop codon-less transcripts, which are produced by endonucleolytic cleavage. We demonstrate that in plants, like in animals, Pelota, Hbs1 and SKI2 proteins are required for NSD, supporting that NSD is an ancient and conserved eukaryotic quality control system. Relevantly, we found that NSD and RNA silencing systems cooperate in plants. Plant silencing predominantly represses target mRNAs through endonucleolytic cleavage in the coding region. Here we show that NSD is required for the elimination of 5' cleavage product of mi- or siRNA-guided silencing complex when the cleavage occurs in the coding region. We also show that NSD and nonsense-mediated decay (NMD) quality control systems operate independently in plants.
DOI: 10.1038/s41598-017-11097-0 |
|
Identification of oriT and a recombination hot spot in the IncA/C plasmid backbone Anna Hegyi, Mónika Szabó, Ferenc Olasz, János Kiss |
A multidrog rezisztenca terjedése a patogén baktériumok között egyre gyorsuló folyamattá vált az elmúlt évtizedekben. A széles gazda-spektrumú konjugatív IncA/C plazmidok fontos szereplői ennek a folyamatnak. A plazmidcsaládnak több, mint 150 tagja ismert, melyek mind több rezisztenciagént is hordoznak, ennek ellenére konjugációs rendszerük, mely révén hatékonyan terjednek a Gram negatív baktériumok körében, még alig ismert. Cikkünkben az IncA/C plazmidok konjugációjának legfontosabb elemét, a transzfer origót (oriT) azonosítjuk és jellemezzük, valamint bizonyítjuk, hogy ez a régió tartalmazza a konjugációhoz elengedhetetlen mobI gén promoterét is. Az oriT szekvencia azonosítása mellett a plazmidon felfedeztünk egy konzervált helyspecifikus rekombinációs rendszert, melynek későbbiekben biotechnológiai alkalmazása is lehetséges.
Dissemination of multiresistance has been accelerating among pathogenic bacteria in recent decades. The broad host-range conjugative plasmids of the IncA/C family are effective vehicles of resistance determinants in Gram-negative bacteria. Although more than 150 family members have been sequenced to date, their conjugation system and other functions encoded by the conserved plasmid backbone have been poorly characterized. The key cis-acting locus, the origin of transfer (oriT), has not yet been unambiguously identified. We present evidence that IncA/C plasmids have a single oriT locus immediately upstream of the mobI gene encoding an indispensable transfer factor. The fully active oriT spans ca. 150-bp AT-rich region overlapping the promoters of mobI and contains multiple inverted and direct repeats. Within this region, the core domain of oriT with reduced but detectable transfer activity was confined to a 70-bp segment containing two inverted repeats and one copy of a 14-bp direct repeat. In addition to oriT, a second locus consisting of a 14-bp imperfect inverted repeat was also identified, which mimicked the function of oriT but which was found to be a recombination site. Recombination between two identical copies of these sites is RecA-independent, requires a plasmid-encoded recombinase and resembles the functioning of dimer-resolution systems.
doi.org/10.3390/genes8120387 |
|
NAD1 Controls Defense-Like Responses in Medicago truncatula Symbiotic Nitrogen Fixing Nodules Following Rhizobial Colonization in a BacA-Independent Manner Ágota Domonkos, Szilárd Kovács, Anikó Gombár, Ernő Kiss, Beatrix Horváth, Gyöngyi Z. Kováts, Attila Farkas, Mónika T. Tóth, Ferhan Ayaydin, Károly Bóka, Lili Fodor, Pascal Ratet, Attila Kereszt, Gabriella Endre, Péter Kaló |
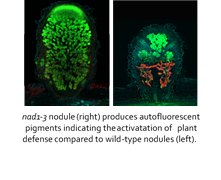
A pillangósvirágú növények és a talajlakó rhizobium baktériumok között létrejövő endoszimbiózis a biológiai nitrogénkötés leghatékonyabb formája. A légköri nitrogén megkötése a növény gyökerén fejlődő speciális szervben, a gyökérgümőben történik, amelyben a növényi sejtekbe bejutó baktériumok átalakulnak nitrogénkötésre képes formájukká, elkezdődik a nitrogénkötés és megindul a szimbiotikus partnerek közötti tápanyagcsere. A gümő kialakulásához elengedhetetlen, hogy a növény a baktériumokat partnerként, és ne betolakodóként ismerje fel. A cikkben egy olyan gén (NAD1) azonosítását és jellemzését írtuk le, amely ahhoz szükséges, hogy a növény patogének ellen irányuló védekező reakcióját kontrolálja a gümőben lehetővé téve a rhizobiumok belépését a növény sejtjeibe és a nitrogénkötésre képes gyökérgümő kialakulását.
Legumes form endosymbiotic interaction with host compatible rhizobia, resulting in the development of nitrogen-fixing root nodules. Within symbiotic nodules, rhizobia are intracellularly accommodated in plant-derived membrane compartments, termed symbiosomes. In mature nodule, the massively colonized cells tolerate the existence of rhizobia without manifestation of visible defense responses, indicating the suppression of plant immunity in the nodule in the favur of the symbiotic partner. Medicago truncatula DNF2 (defective in nitrogen fixation 2) and NAD1 (nodules with activated defense 1) genes are essential for the control of plant defense during the colonization of the nitrogen-fixing nodule and are required for bacteroid persistence. The previously identified nodule-specific NAD1 gene encodes a protein of unknown function. Herein, we present the analysis of novel NAD1 mutant alleles to better understand the function of NAD1 in the repression of immune responses in symbiotic nodules. By exploiting the advantage of plant double and rhizobial mutants defective in establishing nitrogen-fixing symbiotic interaction, we show that NAD1 functions following the release of rhizobia from the infection threads and colonization of nodule cells. The suppression of plant defense is self-dependent of the differentiation status of the rhizobia. The corresponding phenotype of nad1 and dnf2 mutants and the similarity in the induction of defense-associated genes in both mutants suggest that NAD1 and DNF2 operate close together in the same pathway controlling defense responses in symbiotic nodules.
DOI: https://doi.org/10.1016/j.fm.2017.03.011 |
|
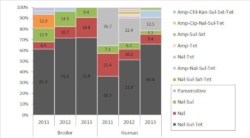
Molecular epidemiology of the endemic multiresistance plasmid pSI54/04 of Salmonella Infantis in broiler and human population Ama Szmolka, Móni Szabó, János Kiss, Judit Pászti, Erzsébet Adrián, |
Salmonella Infantis (SI) became endemic in Hungary where the PFGE cluster B, characterized by a large multiresistance (MDR) plasmid emerged among broilers leading to an increased occurrence in humans. We hypothesized that this plasmid (pSI54/04) assisted dissemination of SI. Indeed, Nal-Sul-Tet phenotypes carrying pSI54/04 occurred increasingly between 2011 and 2013 among SI isolates from broilers and humans. Characterization of pSI54/04 based on genome sequence data of the MDR strain SI54/04 indicated a size of ~277 kb and a high sequence similarity with the megaplasmid pESI of SI predominant in Israel. Molecular characterization of 78 representative broiler and human isolates detected the prototype plasmid pSI54/04 and its variants together with novel plasmid associations within the emerging cluster B. To test in vitro and in vivo pathogenicity of pSI54/04 we produced plasmidic transconjugant of the plasmid-free pre-emergent strain SI69/94. This parental strain and its transconjugant have been tested on chicken embryo fibroblasts (CEFs) and in orally infected day old chicks. The uptake of pSI54/04 did not increase the pathogenicity of the strain SI69/94 in these systems. Thus, dissemination of SI in poultry could be assisted by antimicrobial resistance rather than by virulence modules of the endemic plasmid pSI54/04 in Hungary.
DOI: 10.1099/ijsem.0.002523 |
Xylanibacillus composti gen. nov., sp. nov., isolated from compost
József Kukolya, Ildikó Bata-Vidács Szabina Luzics, Erika Tóth, Zsuzsa Kéki, Peter Schumann, András Táncsics, István Nagy, Ferenc Olasz, Ákos Tóth
A novel Gram-stain-positive bacterial strain, designated as K13T, was isolated from compost and characterized using a polyphasic approach to determine its taxonomic position. On the basis of 16S rRNA gene sequence analysis, the strain showed highest similarity (93.8 %) to Paenibacillus nanensis MX2-3T. Cells of strain K13T were aerobic, motile rods. The major fatty acids were anteiso C15 : 0 (34.4 %), iso C16 : 0 (17.3 %) and C16 : 0 (10.0 %). The major menaquinone was MK-7, the polar lipid profile included diphosphatidylglycerol, phosphatidylethanolamine, phosphatidylglycerol, phosphatidylserine and an aminophospholipid. The DNA G+C content was 52.3 %. Based on phenotypic, including chemotaxonomic characteristics and analysis of the 16S rRNA gene sequences, it was concluded that strain K13T represents a novel genus, for which the name Xylanibacillus gen. nov., sp. nov. is proposed. The type species of the genus is Xylanibacillus composti, the type strain of which is strain K13T (=DSM 29793T=NCAIM B.02605T).
A GENOMIKA FŐOSZTÁLY LEGFRISSEBB PUBLIKÁCIÓI
DOI: doi.org/10.3390/v10060318 |
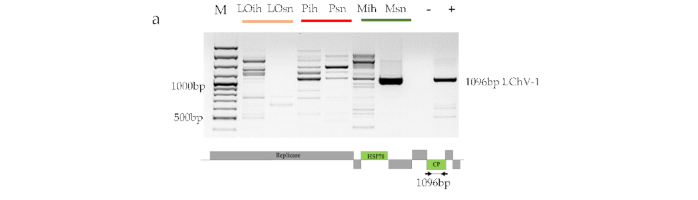
Small RNA NGS Revealed the Presence of Cherry Virus A and Little Cherry Virus 1 on Apricots in Hungary
Baráth, D.; Jaksa-Czotter, N.; Molnár, J.; Varga, T.; Balássy, J.; Szabó, L.K.; Kirilla, Z.; Tusnády, G.E.; Preininger, É.; Várallyay, É.
KisRNS-ek újgenerációs szekvenálásával kajszi ültetvények vírusfertőzöttségét mértük fel. Eredményeink alapján azonosítottuk a cseresznye A vírus és a cseresznye aprógyümölcsűség vírust, utóbbit először ezen a gazdanövényen.
Fruit trees, such as apricot trees, are constantly exposed to the attack of viruses. As they are propagated in a vegetative way, this risk is present not only in the field, where they remain for decades, but also during their propagation. Metagenomic diagnostic methods, based on next generation sequencing (NGS), offer unique possibilities to reveal all the present pathogens in the investigated sample. Using NGS of small RNAs, a special field of these techniques, we tested leaf samples of different varieties of apricot originating from an isolator house or open field stock nursery. As a result, we identified Cherry virus A (CVA) and little cherry virus 1 (LChV-1) for the first time in Hungary. The NGS results were validated by RT-PCR and also by Northern blot in the case of CVA. Cloned and Sanger sequenced viral-specific PCR products enabled us to investigate their phylogenetic relationships. However, since these pathogens have not been described in our country before, their role in symptom development and modification during co-infection with other viruses requires further investigation.
DOI: 10.1007/s00438-017-1412-3 |
|
The red deer Cervus elaphus genome CerEla1.0: sequencing, annotation, genes, chromosomes Bana Á.N., Nyiri A., Nagy J., Frank K., Nagy T., Stéger V., Schiller M., Lakatos P., Sugár L., Horn P., Barta E., Orosz L. |
A Mezőgazdasági Biotechnológiai Központ (MBK,NAIK, Gödöllő), a Kaposvári Egyetem Állattenyésztési, az ELTE Genetikai, és a SOTE 1. Belgy. Klinika MSc és PhD programjai összefogásával, kizárólagosan hazai erőforrásokból, meghatároztatott a gímszarvas DNS-ének szekvenciája és elkészült a gímszarvas teljes genomjának leírása, amely CerELa1.0 névre kereszteltett. A CerEla1.0 a gímszarvas genom első leírása.
We present here the de novo genome assembly CerEla1.0 for the red deer, Cervus elaphus, an emblematic member of the natural megafauna of the Northern Hemisphere. Humans spread the species in the South. Today, the red deer is also a farm-bred animal and is becoming a model animal in biomedical and population studies. Stag DNA was sequenced at 74× coverage by Illumina technology. The ALLPATHS-LG assembly of the reads resulted in 34.7 × 10exp3 scaffolds, 26.1 × 10exp3 of which were utilized in Cer.Ela1.0. The assembly spans 3.4 Gbp. For building the red deer pseudochromosomes, a pre-established genetic map was used for main anchor points. A nearly complete co-linearity was found between the mapmarker sequences of the deer genetic map and the order and orientation of the orthologous sequences in the syntenic bovine regions. Syntenies were also conserved at the in-scaffold level. The cM distances corresponded to 1.34 Mbp uniformly along the deer genome. Chromosomal rearrangements between deer and cattle were demonstrated. 2.8 × 10exp6 SNPs, 365 × 10exp3 indels and 19368 protein-coding genes were identified in CerEla1.0, along with positions for centromerons. CerEla1.0 demonstrates the utilization of dual references, i.e., when a target genome (here C. elaphus) already has a pre-established genetic map, and is combined with the well-established whole genome sequence of a closely related species (here Bos taurus). Genome-wide association studies (GWAS) that CerEla1.0 (NCBI, MKHE00000000) could serve for are discussed.
doi.org/10.3389/fmicb.2018.00122 |
|
NGS of Virus-Derived Small RNAs as a Diagnostic Method Used to Determine Viromes of Hungarian Vineyards N Czotter, J Molnar, E Szabó, E Demian, L Kontra, I Baksa, Gy Szittya, L Kocsis, T Deak, Gy Bisztray, G E. Tusnady, J Burgyan and E Varallyay |
Kis RNS-esk újgenerációs szekvenálásával végeztünk vírusdiagnosztikai felmérést magyar termő szőlőültetvényeken. Amellett, hogy több vírus jelenlétét először igazoltuk hazánkban, eredményeink azt mutatják, hogy az ültetvények vírusfertőzöttségének oka legtöbb esetben a fertőzött szaporítóanyag használata.
As virus diseases cannot be controlled by traditional plant protection methods, the risk of their spread have to be minimized on vegetatively propagated plants, such as grapevine. Metagenomic approaches used for virus diagnostics offer a unique opportunity to reveal the presence of all viral pathogens in the investigated plant, which is why their application can reduce the risk of using infected material for a new plantation. Here we used a special branch, deep sequencing of virus-derived small RNAs, of this high-throughput method for virus diagnostics, and determined viromes of vineyards in Hungary. With NGS of virus-derived small RNAs we could detect not only the viruses tested routinely, but also new ones, which had never been described in Hungary before. Virus presence did not correlate with the age of the plantation, moreover phylogenetic analysis of the identified virus isolates suggests that infections are mostly caused by the use of infected propagating material. Our results, validated by other molecular methods, raised further questions to be answered before this method can be introduced as a routine, reliable test for grapevine virus diagnostics.
DOI: 10.1080/23802359.2017.1390415 |
|
The full mitochondrial genomes of Mangalica pig breeds and their Frank K., Molnár J., Barta E., Marincs F. |
A Mangalica egy régi magyar fajta, melynek megőrzése olyan nemzeti prioritás, amit a Földművelésügyi Minisztérium támogat és szabályoz. Három Mangalica valamint egy Duroc egyed mitokondriális genomszekvenciája került meghatározásra, valamint ezek a teljes mitokondriális szekvenciák összehasonlításra kerültek különböző sertésfajták elérhető mitokondriális genomszekvenciájával.
The mitogenomes of one animal of each of the three Mangalica breeds, Blonde, Red, and Swallow-belly were assembled from reads obtained by Next Generation Sequencing of the three genomes. Features of the mitogenomes were identical in the three breeds, apart from a second tRNA-Val gene on the L strand in Swallow-belly. Phylogenetic comparison of the three mitogenomes with 112 full mtDNA sequences clearly put Mangalicas into the European clade. Comparing the mitogenome of eight Mangalica animals revealed particular differences between them. The mitogenome of some Mangalicas was closely related to the Croatian Turopolje breed and this indicates either the common origin of their maternal lineages or admixture of some populations of the breeds. However, the origin of the mitogenome of certain purebred Mangalicas kept in the Hungarian Mangalica Gene Reserve still remains unknown.
doi.org/10.1007/978-1-4939-7683-6_9 |
|
Use of siRNAs for Diagnosis of Viruses Associated to Woody Plants in Nurseries and Stock Collections Czotter, N., Molnár, J., Pesti, R., Demián, E., Baráth, D., Varga, T., |
A kisRNS-ek újgenerációs szekvenálásának, mint diagnosztikai módszernek lépésről-lépésre történő leírása.
Woody perennial plants like grapevine and fruit trees can be infected by several viruses even as multiple infections. Since they are propagated vegetatively, the phytosanitary status of the propagation material (both the rootstock and the variety) can have a profound effect on the lifetime and health of the new plantations. The fast evolution of sequencing techniques provides a new opportunity for metagenomics-based viral diagnostics. Viral derived small RNAs produced by the host immune system during viral infection can be sequenced by next-generation techniques and analyzed for the presence of viruses, revealing the presence of all known viral pathogens in the sample. This method is based on Illumina sequencing of short RNAs and bioinformatics analysis of virus-derived small RNAs in the host. Here we describe a protocol for this challenging technique step by step with notes, in order to ensure success for every user.
DOI: 10.1016/j.theriogenology.2018.02.021 |
|
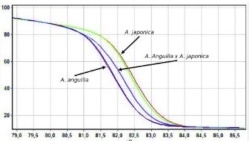
Testing cryopreserved European eel sperm for hybridization Müller T., Matsubara H., Kubara Y., Horváth Á., Kolics B., Taller J., |
A fagyasztás angolna sperma termékenyítőképességére gyakorolt hatásának vizsgálata történt. A hibridek kimutatására PCR-RFLP és PCR-HRM analízis optimalizálás történt.
The objective of this study was to assess impact of cryopreserved European eel sperm and Japanese eel native sperm on early fertilization, hatch, survival, and malformation rates of larvae, as well as develop molecular techniques to distinguish different eel species. Eggs from Japanese eel females (Anguilla japonica) were artificially fertilized with sperm of Japanese eel males and cryopreserved sperm from European eel (A. anguilla, extender was modified Tanaka solution and methanol as cryoprotectant). There were no statistical differences (p > 0.05) among the measured parameters such as fertilization, hatch and survival after 10 days post-hatch rates due to large individual differences. The malformation rate of larvae compared to the hatching rate was higher in cryopreserved groups than in the control indicating that the methodology needs further refinement. Genetic analyses (PCR-RFLP, PCR-HRM) proved a clear result in the detection of paternal contribution in hybridization between the Japanese and the European eel and applied PCR-HRM method is a quick and cost effective tool to identify illegally imported A. anguilla at the glass eel stage, which can be transported from Europe to Asia.
URL: https://www.ornisfennica.org/latest.htm |
|
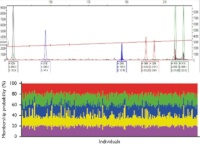
High genetic diversity and weak population structuring in the Schally G., Frank K., Heltai B., Fehér P., Farkas Á., Szemethy L., Stéger V. |
A genetikai monitoring vizsgálatok során egyedi genotipizáláshoz STR (mikroszatellita) markerekből multiplex reakciók lettek összeállítva és optimalizálva. Az egyedi STR profilok egyedi azonosításhoz, valamint a vonuló állományok genetikai diverzitásának és genetikai strukturáltságának vizsgálatához lettek felhasználva.
In this study, we examined the variation of eight autosomal microsatellite markers, and explored the genetic diversity and the measure of population structuring of Eurasian Woodcock (Scolopax rusticola) in Hungary in spring. We tested whether any subpopulations can be identified in our sample, and we also examined whether the individuals that occurred closer to each other in space and in timewere alsomore similar in genetic terms. We analysed samples from 240 free-ranging birds collected during legal hunting from mid-February to the end of April in 2015 from different parts of the country. The Bayesian clustering method andMarkov ChainMonte Carlo simulation were used to infer the most probable number of genetic clusters without a priori definition of populations. The second approach was a discriminant analysis of principal components in order to identify clusters of individuals without population geneticmodels.Additionally,we fitted a general linear model with the genetic distance between samples as the dependent variable, the temporal (days) and the geographical (meters) distances and the interactions of these factors as independent variables.We found high genetic diversity and lowlevel of population structuring in our samples.Moreover, our results did not support the assumptions that Woodcocks occurring in different places or at different times in Hungary would also belong to different breeding populations.
A NÖVÉNYBIOTECHNOLÓGIA FŐOSZTÁLY LEGFRISSEBB PUBLIKÁCIÓI
doi: 10.1371/journal.pone.0200207 |
Expansion of Capsicum annum fruit is linked to dynamic tissue-specific differential expression of miRNA and siRNA profiles.
Taller D, Bálint J, Gyula P, Nagy T, Barta E, Baksa I, Szittya G, Taller J, Havelda Z. PLoS One. 2018 Jul 25;13(7) (IF 2.8)
Szabályozó kis RNS-ek kifejeződésének szintjét vizsgáltuk, szöveti szinten, a paprika termés növekedése során nagy áteresztő képességű szekvenálási módszerrel. Megállapítottuk, hogy a termés növekedés szorosan kapcsolódik egyes kis RNS típusok dinamikus változáshoz, amely az RNS interferencia mechanizmusának fontos szabályozó szerepére utal ebben a gazdasági szempontból is fontos fejlődésbiológiai folyamatban.
Small regulatory RNAs, such as microRNAs (miRNAs) and small interfering RNAs (siRNAs) have emerged as important transcriptional and post-transcriptional regulators controlling a wide variety of physiological processes including fruit development. Data are, however, limited for their potential roles in developmental processes determining economically important traits of crops. The current study aimed to discover and characterize differentially expressed miRNAs and siRNAs in sweet pepper (Capsicum annuum) during fruit expansion. High-throughput sequencing was employed to determine the small regulatory RNA expression profiles in various fruit tissues, such as placenta, seed, and flesh at 28 and 40 days after anthesis. Comparative differential expression analyses of conserved, already described and our newly predicted pepper-specific miRNAs revealed that fruit expansion is accompanied by an increasing level of miRNA-mediated regulation of gene expression. Accordingly, ARGONAUTE1 protein, the primary executor of miRNA-mediated regulation, continuously accumulated to an extremely high level in the flesh. We also identified numerous pepper-specific, heterochromatin-associated 24-nt siRNAs (hetsiRNAs) which were extremely abundant in the seeds, as well as 21-nt and 24-nt phased siRNAs (phasiRNAs) that were expressed mainly in the placenta and the seeds. This work provides comprehensive tissue-specific miRNA and siRNA expression landscape for a developing pepper fruit. We identified several novel, abundantly expressing tissue- and pepper-specific small regulatory RNA species. Our data show that fruit expansion is associated with extensive changes in sRNA abundance, raising the possibility that manipulation of sRNA pathways may be employed to improve the quality and quantity of the pepper fruit.
DOI: 10.1111/pbi.13077. |
Creating highly efficient resistance against wheat dwarf virus in barley by employing CRISPR/Cas9 system.
Kis A, Hamar É, Tholt G, Bán R, Plant Havelda Z. Biotechnol J. 2019 Jan 11. (IF 6.3)
A CRISPR/ Cas9 alapú genom szerkesztési módszert használtuk fel közvetlenül egy DNS vírus, Wheat dwarf virus (WDV), inaktiválására árpa növényben. Eredményeink megmutatták, hogy ez az eljárás alkalmas egy gazdasági szempontból kiemelt fontosságú korokozó ellen teljes rezisztenciát illetve nagyfokú toleranciát kialakítani egyszikű haszonnövényben.
Wheat dwarf virus (WDV) is an economically important, insect-transmitted virus belonging to the Geminiviridae family. WDV strains infect both wheat and barley causing severe yield losses, and the natural resistance resources are limited. Direct utilization of the CRISPR/Cas9 system to inhibit geminivirus replication has been described in model plants. Here we show the direct anti-viral utilization of the CRISPR/Ca9 system in an important crop plant, barley (Hordeum vulgare L. cv. Golden promise), to establish an effective WDV resistance. Artificial transient expression system has been used to test the biological activity of selected WDV-specific single guide RNAs (sgRNAs). Stable introduction of multiple-gene cassette expressing four WDV-specific sgRNAs and Cas9 into barley plants resulted in an extreme resistance in the majority of the challenge infection experiments mediated by virus-carrying leafhoppers. The introduced resistance was stably heritable providing resistance to the progeny lines. However, we also found that in a few cases WDV escaped from the control of the CRISPR/Cas9 system. Molecular analyses of the infected barley lines revealed that only one sgRNA was active probably imposing high but in some cases surmountable selection pressure on the replicating virus. Our results show that the CRISPR/Cas9-mediated direct virus targeting can be a powerful tool to engineer full resistance against economically important, phloem-limited, geminiviruses in monocotyledonary crop plants, but the design of multiple, biologically highly active sgRNAs is the prerequisite of its durable utilization.
DOI: https://doi.org/10.1111/pce.13355 |
|
Ambient temperature regulates the expression of a small set of sRNAs Gyula P, Baksa I, Tóth T, Mohorianu I, Dalmay T, Szittya G. |
Our previous study on temperature-dependent antiviral silencing suggested that the endogenous silencing regulating plant development is also under temperature control. Surprisingly, the comparison of sRNA populations at 15, 21 and 27 °C in Arabidopsis thaliana seedlings, roots, leaves, and flowers revealed that only a small fraction of the endogenous small RNAs is temperature-regulated. We found that miR169, targeting NF-YA2 and JAZ4, and a hc-siRNA locus regulating YUC2 might have a role in temperature adaptation.
Plants substantially alter their developmental program upon changes in the ambient temperature. The 21–24 nt small RNAs (sRNAs) are important gene expression regulators, which play a major role in development and adaptation. However, little is known about how the different sRNA classes respond to changes in the ambient temperature. We profiled the sRNA populations in four different tissues of Arabidopsis thaliana plants grown at 15, 21 and 27 °C. We found that only a small fraction (0.6%) of the sRNA loci are ambient temperature‐controlled. We identified thermoresponsive miRNAs and identified their target genes using degradome libraries. We verified that the target of the thermoregulated miR169, NF‐YA2, is also ambient temperature‐regulated. NF‐YA2, as the component of the conserved transcriptional regulator NF‐Y complex, binds the promoter of the flowering time regulator FT and the auxin biosynthesis gene YUC2. Other differentially expressed loci include thermoresponsive phased siRNA loci that target various auxin pathway genes and tRNA fragments. Furthermore, a temperature dependent 24‐nt heterochromatic siRNA locus in the promoter of YUC2 may contribute to the epigenetic regulation of auxin homeostasis. This holistic approach facilitated a better understanding of the role of different sRNA classes in ambient temperature adaptation of plants.
DOI: 10.1038/s41598-017-01050-6 |
|
Crispr/Cas9 Mediated Inactivation of Argonaute 2 Reveals its Differential Involvement in Antiviral Responses Márta Ludman, József Burgyán & Károly Fátyol |
RNA silencing constitutes an important antiviral mechanism in plants. Small RNA guided Argonaute proteins fulfill essential role in this process by acting as executors of viral restriction. Plants encode multiple Argonaute proteins of which several exhibit antiviral activities. A recent addition to this group is AGO2. Its involvement in antiviral responses is established predominantly by studies employing mutants of Arabidopsis thaliana. In the virological model plant, Nicotiana benthamiana, the contribution of AGO2 to antiviral immunity is much less certain due to the lack of appropriate genetic mutants. Previous studies employed various RNAi based tools to down-regulate AGO2 expression. However, these techniques have several disadvantages, especially in the context of antiviral RNA silencing. Here, we have utilized the CRISPR/Cas9 technology to inactivate the AGO2 gene of N. benthamiana. The ago2 plants exhibit differential sensitivities towards various viruses. AGO2 is a critical component of the plants' immune responses against PVX, TuMV and TCV. In contrast, AGO2 deficiency does not significantly influence the progression of tombusvirus and CMV infections. In summary, our work provides unequivocal proof for the virus-specific antiviral role of AGO2 in a plant species other than A. thaliana for the first time.
Programajánló
Hírek
Tisztelt Látogatók!
A hazai agrár-felsőoktatás szükséges megújulásának mérföldköve az alapítványi fenntartású Magyar Agrár- és Élettudományi Egyetem (MATE) létrejötte, amely 2021. február 1-től 5 campuson, több mint 13 ezer hallgató számára fogja össze a dunántúli és közép-magyarországi élettudományi és kapcsolódó képzéseket. Az intézményhez csatlakozik a Nemzeti Agrárkutatási és Innovációs Központ (NAIK) 11 kutatóintézete is, így az új intézmény nem csupán egy oktatási intézmény lesz, hanem az ágazat szellemi, szakpolitikai és innovációs központjává válik, amely nagyobb mozgásteret biztosít a képzések, a gazdálkodás és szervezet modernizálásához, fejlesztéséhez. Az összeolvadással magasabb fokozatra kapcsolunk, a kutatói és egyetemi szféra szorosabban fonódik majd össze, aminek következtében még több érdekes, izgalmas kutatás-fejlesztés születhet majd az agrárium területén.
Kérjük, kövesse tevékenységünket a jövőben is a www.uni-mate.hu honlapon!
2021. február 1-től a Nemzeti Agrárkutatási és Innovációs Központ 11 kutatóintézete, köztük az MBK is a Szent István Egyetemmel együtt Magyar Agrár- és Élettudományi Egyetem néven, közhasznú magán felsőoktatási intézményként,átalakított szervezeti struktúrával folytatja tovább működését.