ABC - Department for Genetics - Laboratory of Microbiology
Current Research Projects
- Isolation and identification of microorganisms with biotechnological importance and their characterisation by microbiological and molecular biological approaches
- Development of new „starter” cultures, highlighting the character of wine from Balaton wine region
- Isolation and characterization of yeast strains from spontaneously fermented wines
Isolation and identification of microorganisms with biotechnological importance and their characterisation by microbiological and molecular biological approaches
The microorganisms present in the gastrointestinal tract of animals form the so called gut microbiota, whose quantitative and qualitative balance is an important factor in the health of the concerned animals. The disruption of this balance may cause the development of various diseases; however, some microorganisms may also facilitate the stabilization of a “balanced” microbiota, thus contributing to a recovery process. In industrial livestock production the animals often suffer from health problems whose alleviation or prevention may be achieved by using a so called „probiotic” product, developed for the concerned species.
Probiotic properties are strain specific, and probiotics must comply with a number of criteria. Therefore, an important part of the work is the identification and characterisation of the isolates at the species and strain levels. The experiments involve the sampling of the gastrointestinal content from various sources, the isolation of primarily bacterial strains and their maintenance in a strain collection. Our molecular biological approach is essentially PCR based. The microbiological characterisation of the strains covers their antibacterial properties and antibiotic susceptibility, moreover, beyond the purpose-oriented selection the testing also includes whole genome sequencing and molecular marker development for selected isolates.
In addition to the applied research activities described above, we are also involved in basic research through microbiological profiling and metagenomic analyses of the microbiota of various biological samples that are planned to be presented at conferences and in scientific papers.
Development of new „starter” cultures, highlighting the character of wine from Balaton wine region
The utilization of „starter” yeast is very important strategy for controlling the fermentation in modern wine-making. Applying of these strains avoid the risk of slow or incomplete fermentation. However, the use of “commercial” strains can results in uniform or similar characteristic of wines. Lot of starter yeast products are available in trade-market originating usually from natural population. Different wine region have unique microbiom and these microbiom acclimated to the local environmental condition. Some of them are playing roll in indigenous wine-making (spontaneous fermentation) and secondary metabolite production of these yeasts influence the character of wine.
Our aim is isolation of yeast strains from Badacsony vineyards and characterize with molecular, microbial and genomic methods. Our perspective goal is the collected indigenous yeast strains that could be implemented in the development of “terroir” starter cultures to be used in the production of “organic wines”.
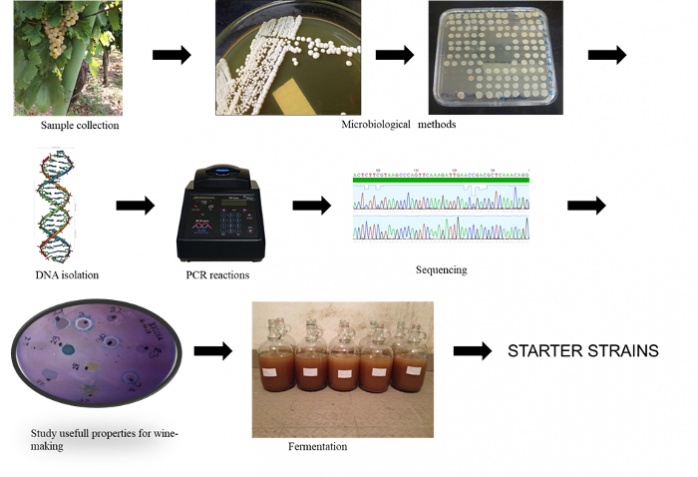
Isolation and characterization of yeast strains from spontaneously fermented wines
The growing demand for wines with individual character and typical regional features has led to the study of indigenous yeast strains. Due to the determinant role that the yeast strain plays as regards to wine quality, the terroir concept includes not only the soil, climate or weather in a region as major aspects, but also the indigenous yeast strains.
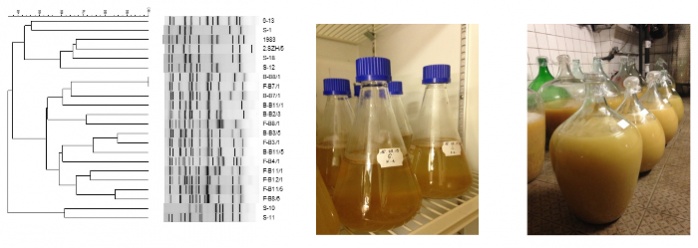
The aim of our research is to isolate indigenous wine yeast strains from the Hungarian wine regions and investigate their potential application as starter culture. For the identification and characterization of the isolated strains, we use molecular biology methods (Saccharomyces genus specific PCR, mtDNA-RFLP, interdelta analysis). Microbiological characteristics (ethanol-, sulfur dioxide-, osmo-tolerance, acid, and hydrogen sulfide production, killer activity) of the S. cerevisiae isolates are also studied. The evaluation of the oenological potential of the isolates is carried out in micro- and mesovinification tests. The analytical and sensory properties of the mesovinification products (wines) are investigated and compared. In the future, we are planning to explore the microbiota of the spontaneously fermented wines by using metagenomic analysis.
Current Research Projects
Enquiries from MSc and potential PhD students are very welcomed. Please feel free to contact us by email (olasz.ferenc@abc.naik.hu), if interested.
Laboratory of Microbiology
Staff
Publications
Selected Publications
Anna Hegyi, Mónika Szabó, Ferenc Olasz, János Kiss (2017)
Identification of oriT and a recombination hot spot in IncA/C plasmid backbone
Scientific Reports 2017 Sep 6;7(1):10595. doi: 10.1038/s41598-017-11097-0.
Német Z, Albert E, Nagy T, Olasz F, Barta E, Kiss J, Dán Á, Bányai K, Hermans K, Biksi I. 2017.
Draft genome sequence of an atypical highly virulent rabbit Staphylococcus aureus strain.
Genome Announc 5:e01049-17. https://doi.org/10.1128/genomeA.01049-17.
Veress A, Wilk T, Kiss J, Papp PP, Olasz F. (2017) Two Draft Genome Sequences of Sphingobacterium sp. Strains Isolated from Honey.
Genome Announc. 2017 Nov 30;5(48). pii: e01364-17. doi: 10.1128/genomeA.01364-17.
József Kukolya, Ildikó Bata-Vidács, Szabina Luzics, Erika Tóth, Zsuzsa Kéki, Peter Schumann, András Táncsics, István Nagy, Ferenc Olasz, Ákos Tóth (2018)
Xylanibacillus composti gen. nov., sp. nov., isolated from compost
International Journal of Systematic and Evolutionary Microbiology 68: 698-702, doi: 10.1099/ijsem.0.002523
Ama Szmolka, Móni Szabó, János Kiss, Judit Pászti, Erzsébet Adrián, Ferenc Olasz, Béla Nagy (2017)
Molecular epidemiology of the endemic multiresistance plasmid pSI54/04 of Salmonella Infantis in broiler and human population in Hungary.
Food Microbiology, in press
Wilk T, Szabó M, Szmolka A, Kiss J, Olasz F, Nagy B. (2017)
Genome Sequences of Salmonella enterica subsp. enterica Serovar Infantis Strains from Hungary Representing Two Peak Incidence Periods in Three Decades.
Genome Announc. 2017 Mar 2;5(9). pii: e01735-16. doi: 10.1128/genomeA.01735-16.
Á. Tóth , E. Baka , Sz. Luzics , I. Bata-Vidács , I. Nagy , B. Bálint , R. Herczeg , F. Olasz, T. Wilk , T. Nagy , B. Kriszt , I. Nagy , J. Kukolya(2016)
Plant polysaccharide degrading enzyme system of Thermobifida cellulosilytica TB100T revealed by de novo genome project dataActa AlimentariaPosted online on 31 Mar 2016
Wilk T, Szabó M, Szmolka A, Kiss J, Barta E, Nagy T, Olasz F, Nagy B.
Genome Sequences of Multidrug-Resistant Salmonella enterica subsp. enterica Serovar Infantis Strains from Broiler Chicks in Hungary.
Genome Announc. 2016 Dec 15;4(6). pii: e01400-16. doi: 10.1128/genomeA.01400-16
Gábor Murányi, Mónika Szabó, Ferenc Olasz, János Kiss (2016)
Determination and analysis of the putative AcaCD-responsive promoters of SGI1
PLoS ONE 11(10): e0164561. doi:10.1371/journal.pone.0164561
Mónika Szabó, Tibor Nagy,Tímea Wilk, Tibor Farkas, Anna Hegyi,Ferenc Olasz, János Kiss.
Characterization of two multidrug-resistant IncA/C plasmids from the 1960s by using Oxford Nanopore MinION sequencer device.
Antimicrob Agents Chemother. 2016 Sep 6. pii: AAC.01121-16. [Epub ahead of print]
Ariel Imre, Ama Szmolka, Ferenc Olasz, Béla Nagy (2015)
Vaccine potential of a nonflagellated virulence-plasmid cured, (fliD-, pSEV∆) mutant of Salmonella Enteritidis for chickens.
Acta Veterinaria Hungarica 09/2015; 63(3):285-302..
János Kiss*, Péter Pál Papp, Mónika Szabó, Tibor Farkas, Gábor Murányi, Erik Szakállas and Ferenc Olasz* (2015)
The master regulator of IncA/C plasmids is recognized by the Salmonella Genomic island SGI1 as a signal for excision and conjugal transfer
Nucleic Acid Reserach 2015 Oct 15;43(18):8735-45. doi: 10.1093/nar/gkv758. Epub 2015 Jul 24
Német Z, Albert E, Nagy T, Olasz F, Barta E, Kiss J, Dán Á, Bányai K, Hermans K, Biksi I.
Draft Genome Sequence of a Highly Virulent Rabbit Staphylococcus aureus Strain.
Genome Announc. 2015 Jul 9;3(4). pii: e00461-15. doi: 10.1128/genomeA.00461-15.
Ferenc Olasz,a#* Tibor Nagy,a* Móni Szabó,a János Kiss,a Ama Szmolka,b Endre Barta,a Andries van Tonder,c Nicholas Thomson,d Paul Barrow,e# Béla Nagyb (2015)
Genome Sequences of three Salmonella enterica subsp.enterica serovar Infantis strains from healthy broiler chicks in Hungary and in the United Kingdom. Genome Announcements 3(1):e01468-14. doi:10.1128/genomeA.01468-14
Olasz F., Nagy T., Szabó M., Kiss J., Szmolka A., Barta E., van Tonder A., Thomson N., Barrow P., Nagy B. (2015).
Genome Sequences of Three Salmonella enterica subsp. enterica Serovar Infantis Strains from Healthy Broiler Chicks in Hungary and in the United Kingdom. Genome Announc. 3(1). pii: e01468-14. doi: 10.1128/genomeA.01468-14.
Kiss, János, Béla Nagy, Ferenc Olasz (2012).
Stability, entrapment and variant formation of Salmonella genomic island 1.
PLo S ONE 7(2): e32497.
Fekete Péter Z., Elzbieta Brzuszkiewicz, Gabriele Blum-Oehler, Ferenc Olasz, Mónika Szabó, Gerhard Gottschalk, Jörg H. Hacker and Béla Nagy (2012). Sequence analysis and comparative pathogenomics of plasmid pTc conferring virulence and antimicrobial resistance for F18+ porcine enterotoxigenic Escherichia coli.
Int. J. Med. Microbiol. 302, 4-9.
Ariel Imre, Ferenc Olasz, Béla Nagy (2011).
Site-directed (IS30-FljA) transposon mutagenesis system to produce non-flagellated mutants of Salmonella enteritidis.
FEMS Microbiol. Lett. 317, 52–59.
Szabó M., Kiss J. and Olasz F.(2010):
Functional organization of the inverted repeats of IS30.
J. Bacteriol. 192, 3414-23.
Deak, V., Lukacs, R., Buzas, Z., Palvölgyi, A., Papp, P. P., Orosz, L. and Putnoky, P. (2010).
Identification of tail genes in the temperate phage 16-3 of Sinorhizobium meliloti 41.
J. Bacteriol. 192,1617-1623.
Szabó M., Kiss J., Nagy Z.,Chandler M. and Olasz F. (2008).
Sub-terminal sequences modulating IS30 transposition in vivo and in vitro.
J.Mol.Biol. 375, 337-52.
Bodogai, M., Ferenczi, Sz., Miclea, S. P., Papp, P. and Dusha, I. (2008). Toxin-amtitoxin modules and symbiosis. In “Biological Nitrogen Fixation: Towards Poverty Alleviation Through Sustainable Agriculture” (Dakora, F.D., Chimphango, S.B.M., Valentine, A.J. Elmerich, C. and Newton, W.E., eds), Book Series: Current Plant Science and Biotechnology in Agriculture 42, pp. 237-238.
Ferg M, Sanges R, Gehrig J, Kiss J, Bauer M, Lovas A, Szabo M, Yang L, Straehle U, Pankratz MJ, Olasz F, Stupka E, Müller F. (2007).
The TATA-binding protein regulates maternal mRNA degradation and differential zygotic transcription in zebrafish.
EMBO J. 26, 3945-56.
Kiss János, Zita Nagy,Gábor Tóth, György Botond Kiss, Júlia Jakab, Michael Chandler, Ferenc Olasz (2007).
Transposition and target specificity of the typical IS30 family element IS1655 from Neisseria meningitidis.
Molec. Microbiol. 63, 1731-1747.
Ferenczi, S., Orosz, L. and Papp, P. P. (2006).
Repressor of 16-3 Phage with Altered Binding Specificity Indicates Spatial Differences in Repressor-Operator Complexes.
J. Bacteriol. 188, 1663-1666.
Das, N., Valjavec-Gratian, M., Basuray, A. N., Fekete, R. A., Papp, P. P., Paulsson, J. and Chattoraj, D. K. (2005).
Multiple homeostatic mechanisms in the control of P1 plasmid replication.
Proc. Natl. Acad. Sci. U.S.A. 102, 2856-2861.
Ganyu, A., Csiszovszki, Z., Ponyi, T., Kern, A., Buzas, Z., Orosz, L. and Papp, P. P. (2005).
Identification of cohesive ends and genes encoding the terminase of phage 16-3.
J. Bacteriol. 187, 2526-2531.
Nagy Zita, Mónika Szabó,Michael Chandler and Ferenc Olasz (2004).
Analysis of the N-terminal DNA binding domain of the IS30 transposase.
Molec. Microbiol. 54, 478-88.
Kiss János, Mónika Szabó and Ferenc Olasz(2003):
Site-specific recombination by the DDE-family member IS30 transposase.
Proc Natl Acad Sci U S A. 100, 1500-05.
Olasz Ferenc, Tamás Fischer, Mónika Szabó, Zita Nagy and János Kiss (2003).:
Gene conversion in transposition of Escherichia coli element IS30.
J. Mol. Biol. 334, 967-978.
Fekete Péter Zsolt, György Schneider, Ferenc Olasz, Gabriele Blum-Oehler, Jörg H. Hacker, Béla Nagy (2003):
Detection of a plasmid encoded pathogenicity island in F18+ enterotoxigenic and verotoxigenic Escherichia coli from weaned pigs.
Int. J. Med. Microbiol. 293, 287-298.
Szabó Mónika, Ferenc Müller, János Kiss, Carolin Balduf, Uwe Strähle and Ferenc Olasz (2003).
Trans-kingdom transposition and gene targeting mediated by the prokaryotic mobile element IS30.
FEBS Lett. 550, 46-50.
Nagy, Z., Szeverényi, I.,Farkas, T., Olasz, F. and Kiss J.(2003).
Detection and analysis of transpositionally active head-to-tail dimers in three additional E. coli IS elements.
Microbiol. 149, 1297-1310.
Papp, P. P.,Nagy, T., Ferenczi, S., Elo, P., Csiszovszki, Z., Buzas, Z., Patthy, A. and Orosz, L. (2002).
Binding sites of different geometries for the 16- 3 phage repressor.
Proc. Natl. Acad. Sci. U.S.A. 99, 8790-8795
Patent
Nagy Béla (50%), Olasz Ferenc (30%) Fekete Péter Zsolt (15%) Tóthné, Szekrényi Márta (5%) (1999). Élő, szájon át adható Escherichia coli vakcina készítésére alkalmas törzs a sertések választási hasmenésének megelőzésére és a törzs előállítására alkalmas eljárás. Magyar Szabadalmi Hivatal, iktatószám: 12917/99, ügyszám: P99 00836.Benyújtó: MTA Állatorvostud. Kutató.
Mónika Szabó, János Kiss, Ferenc Olasz, (ABC, Gödöllő, Hungary), Ferenc Müller, Lászlo Tora, Uwe Strähle (IGBMC, Strasbourg, France). European Patent application, No 01402754.4. Site-directed recombinase fusion proteins and corresponding polynucleotides, vectors and kits, and their uses for site-directed DNA recombination.
Nagy Béla, Olasz Ferenc, Fekete Péter Zsolt (2007). Escherichia coli strain for an oral vaccine against post-weaning diarrhea in pigs.United States Patent US 7,163820 B1.