ABC - Department for Genetics - Host-pathogen interaction and microbial genomics group
Research Interests
The rapid evolution of bacteria due to their outstanding genome plasticity represents a serious challenge for researchers as well as for actors in agriculture or human and animal healthcare. Genomic islands, plasmids and other mobile genetic elements play crucial role in spreading the pathogenicity factors and antibiotic resistance, thus in rapid adaptation and evolution of bacteria. Our aim is to detect and analyze these changes both at molecular and genomic levels. We pay special attention to the molecular background and regulation of horizontal transfer of the Salmonella genomic island 1 (SGI1) causing multiresistance in many Salmonella serovars and other pathogenic bacteria such as Proteus, Acinetobacter and Morganella and to the mechanisms of cooperation between SGI1 and the multiresistance IncC family plasmids that are responsible for horizontal transfer of the island. Our work covers also the investigations of a non-invasive but highly successful Salmonella serovar Infantis, which became prevalent in the last several years and may become the startpoint of a potential pathogen clone in the future. Based on our research our aim is to develop bacterial strains that can be applied in the agriculture, animal healthcare or environmental protection.
Current Research Projects
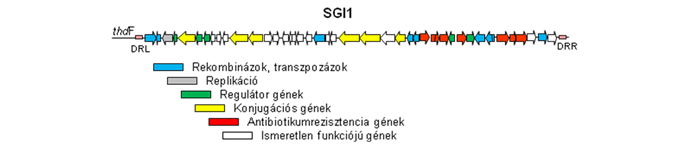
- Genetics, evolution and horizontal transfer of Salmonella genomic island 1.
- Test program of the third generation sequencing system MinION
- Transposition and the emergence of antibiotic resistance in bacteria
- Genomics, evolution and pathogenetic profile of emerging multidrug-resistant Salmonella Infantis strains
Genetics, evolution and horizontal transfer of Salmonella genomic island 1.
The genomic and pathogenicity islands represent considerable part of the accessory genomes of bacteria. Due to their mobility and highly variable genetic structure, these islands significantly contribute to the plasticity of bacterial genomes, thus they are often responsible for the rapid evolution of their host organism.
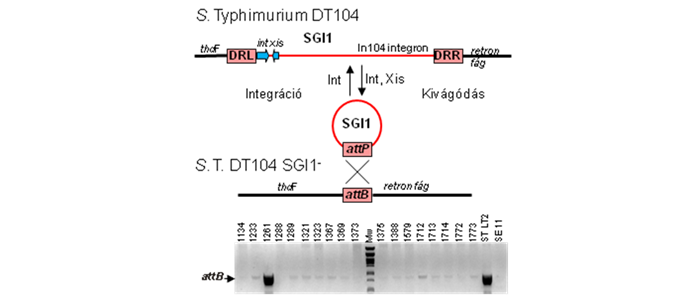
The Salmonella genomic island 1 identified in multiresistant Salmonella Typhimurium strains in early 1990s appears to be a good model for research on genomic islands. SGI1 and its numerous variants are now widely distributed among several epidemic Salmonella serovars, Proteus mirabilis, Acinetobacter baumannii and Morganella morganii strains and represent significant health risk for both humans and animal healthcare. Despite of these facts, molecular mechanisms of SGI1 distribution and the SGI1-host interactions are mainly unknown.
Our work is focused on the discovery of SGI1 genetics with special attention to the mechanisms of horizontal transfer of the island (site-specific recombination, conjugation), the aspects of cooperation with IncC group plasmids involved in the mobilization of the island, and the possible origin and evolution (variant formation) of SGI1. We are also examine the relations of the island and its host organism, especially regarding the role of SGI1 in the pathogenicity and fitness of Salmonella and the in vivo transfer of SGI1 in the gut flora of host animal. In this project our collaborators are the Enteric Bacteriology and Foodborne Zoonoses Group of the Institute for Veterinary Medical Research, Centre for Agricultural Research, Hungarian Academy of Sciences and the Genomic Plasticity, biodiversity and Antimicrobial Resistance Group of INRA-Université de Tours Infectiologie & Santé Publique, Tours, France.
Test program of the third generation sequencing system MinION
Since 2014 our group has participated the worldwide test program MinIon Access Program (MAP) of the third generation sequencing device MinIon developed by Oxford Nanopore Technologies Ltd. The MinION sequencer device is similar in size to a USB memory stick and connected to a PC through USB port. The MinION consists of the device and a consumable flow cell. The flow cell has a sensor chip containing 512 microwells, each one supporting an individual protein nanopore sensor. Each well is a single addressable electronic channel and each nanopore is capable of individual identification of analyte molecules. DNA sequence is determined according to the changes in ion flow through the nanopore during basecalling process. One of the advantages of this technology is that very long (50-100 up to 2000 kb) reads can be generated which makes the de novo sequence assembly much easier.
Source: Oxford Nanopore, https://www.nanoporetech.com/
In the frame of MAP, we have sequenced several multiresistant IncC family plasmids that are ~160-170 kb single copy conjugative plasmids harbouring many different IS elements, which make almost impossible to order the contigs obtained from de novo assembly of short (3-400 bp) reads generated by NGS platforms. In the first part of MAP we applied the long reads produced by MinIon in determination of correct sequence of these plasmids even though their extensive repetitive regions. We hope that further improvements of the system will allow its common application in our genomics studies.
Transposition and the emergence of antibiotic resistance in bacteria
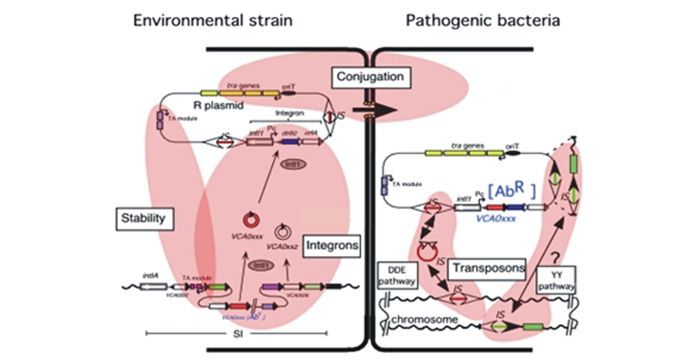
Insertion sequences (IS) are mobile DNA segments possessing the ability to translocate or even mediate large scale rearrangements in the genome of host organisms. These mobile elements play a crucial role in horizontal gene transfer, disseminating for example antibiotic resistance (AR) genes among bacteria due to their ability to cause rearrangements between the chromosome and transmissible genetic elements that are efficient vehicles of the acquired genes even between species. AR represents a serious problem in therapy since the beginning of antibiotic era. One of the most important driving forces behind the rapid acquisition of AR genes by pathogens is that insertion sequences can capture AR genes in various ways to form transposons, which are subsequently translocated onto conjugative plasmids or bacteriophages, which can be transferred with high frequency among bacteria.
A scheme of the various mechanisms of AR gene transmission (the figure is based on a EU CRAB project): movement and exchange of integron cassettes, transposition mechanisms, conjugal transfer between bacteria and fixation of the acquired AR genes.
Using in vivo and in vitro approaches we have determined that the E. coli element IS30 has adopted a transposition mechanism, which involves the formation of a transposon circle that is subsequently integrates into the target DNA. The IR-IR junction of the transposon circle is recombinationally active in the presence of transposase, and generates insertions, deletions inversions or replicon fusions. This type of copy-paste mechanism now appears to be the most widespread among bacterial ISs. Since the circle formation process involves the resolution of a cruciform structure containing free DNA ends, this may induce different repair mechanisms. Our further aim is to explore the interactions between the transposon and its host organism by microarray analysis, network analysis and experimental approaches.
Genomics, evolution and pathogenetic profile of emerging multidrug-resistant Salmonella Infantis strains (in collaboration with the Enteric Bacteriology and Foodborne Zoonoses Group of the Institute for Veterinary Medical Research, Centre for Agricultural Research, Hungarian Academy of Sciences).
Numerous Salmonella serovars represent a serious risk both in human and veterinary medicine. Comparing recent S. Infantis isolates to those from the 80’s striking differences in the occurrence and distribution of antibiotic resistance were detected, indicating the rapid evolution of S. Infantis. Our aim is to study the genetic background of the rapid and probably clonal spread of the multiresistant S. Infantis strains in Hungary and worldwide. Based on the accumulation of whole genome sequencing data, genomes of many ancient and recently emerged isolates can be compared. We intend to answer what kind of genetic rearrangements can explain the rapid spread of Infantis, e.g. what kind of new plasmids or genomic/pathogenicity islands have been acquired or what changes (gain or loss of transposons, integrons) occurred in the genome of the recently evolved strains. To examine the large scale genomic changes we have developed tools for genome manipulations that can help to answer these issues. This research would help to understand the evolution of multidrug resistant clones and the reasons of their success and rapid distribution.
Bacteria on the surface of villi in the intestine. (Dr Béla Nagy, MTA ATK ÁOTI)
Our work is supported by:
NKFIH K 128203 Interrelation of different mobile multiresistance factors: Crosstalk between IncA/C plasmids and SGI1-family genomic islands. (2018-2022).
Hungarian Scientific Research Found OTKA
K 105635 Molecular genetics and ecology of Salmonella Genomic Island 1 (SGI1): secrets of mobility, spread and pathogenetic significance (2012-2017)
K 101546 Evolution and pathogenetic profile of emerging multidrug-resistant Salmonella Infantis (2012-2016)
Staff

Publications
Selected publications
Szmolka A, Szabó M, Kiss J, Pászti J, Adrián E, Olasz F, Nagy B (2018) Molecular epidemiology of the endemic multiresistance plasmid pSI54/04 of Salmonella Infantis in broiler and human population in Hungary. Food Microbiol. 71:25-31. doi: 10.1016/j.fm.2017.03.011. http://dx.doi.org/10.1016/j.fm.2017.03.011.
Anna Hegyi, Mónika Szabó, Ferenc Olasz, János Kiss.(2017) Identification of oriT and a recombination hot spot in the IncA/C plasmid backbone. Scientific Reports, Sci Rep. 6;7(1):10595. doi: 10.1038/s41598-017-11097-0
Veress A., Wilk T., Kiss J., Papp P. P., Olasz F. (2017) Two Draft Genome Sequences of Sphingobacterium sp. Strains Isolated from Honey. Genome Announc. 5(48). pii: e01364-17. doi::10.1128/genomeA.01364-17.
Wilk T, Szabó M, Szmolka A, Kiss J, Olasz F, and Nagy B (2017) Genome Sequences of Salmonella enterica subsp. enterica Serovar Infantis Strains from Hungary Representing Two Peak Incidence Periods in Three Decades. Genome Announc. 5: e01735-16.doi:10.1128/genomeA.01735-16
Mónika Szabó, Tibor Nagy, Tímea Wilk, Tibor Farkas, Anna Hegyi, Ferenc Olasz, János Kiss (2016) Characterization of Two Multidrug-Resistant IncA/C Plasmids from the 1960s by Using the MinION Sequencer Device. Antimicrob Agents Chemother 60:6780–6786. doi:10.1128/AAC.01121-16.
Gábor Murányi, Mónika Szabó, Ferenc Olasz, János Kiss (2016) Determination and Analysis of the Putative AcaCD-Responsive Promoters of Salmonella Genomic Island 1. PLoS ONE 11(10): e0164561. doi:10.1371/journal.pone.0164561
Wilk T, Szabó M, Szmolka A, Kiss J, Barta E, Nagy T, Olasz F, Nagy B. (2016) Genome Sequences of Multidrug-Resistant Salmonella enterica subsp. enterica Serovar Infantis Strains from Broiler Chicks in Hungary. Genome Announc. 4(6). pii: e01400-16. doi: 10.1128/genomeA.01400-16.
Kiss, J., Papp, P., P., Szabó, M., Farkas, T., Murányi, G., Szakállas E. and Olasz F. (2015) The master regulator of IncA/C plasmids is recognized by the Salmonella Genomic island SGI1 as a signal for excision and conjugal transfer. Nucleic Acids Res. 43 (18): 8735-8745 doi:10.1093/nar/gkv758
Olasz F., Nagy T., Szabó M., Kiss J., Szmolka A., Barta E., van Tonder A., Thomson N., Barrow P., Nagy B. (2015) Genome Sequences of Three Salmonella enterica subsp. enterica Serovar Infantis Strains from Healthy Broiler Chicks in Hungary and in the United Kingdom. Genome Announc. 3(1). pii: e01468-14. doi: 10.1128/genomeA.01468-14.
Kiss, János,Béla Nagy, Ferenc Olasz,(2012) Stability, entrapment and variant formation of Salmonella genomic island 1. PLo S ONE 7(2): e32497.
Fekete Péter Z., Elzbieta Brzuszkiewicz, Gabriele Blum-Oehler, Ferenc Olasz, Mónika Szabó,Gerhard Gottschalk, Jörg H. Hacker and Béla Nagy (2012) Sequence analysis and comparative pathogenomics of plasmid pTc conferring virulence and antimicrobial resistance for F18+ porcine enterotoxigenic Escherichia coli. Int J Med Microbiol. 2011 Oct 12. [Epub ahead of print] 302, 4-9.
Szabó M., Kiss J., and Olasz F.(2010): Functional organization of the inverted repeats of IS30. J Bacteriol. 2010 Jul;192(13):3414-23. Epub 2010 Apr 23.
Szabó M., Kiss J., Nagy Z.,Chandler M. and Olasz F. (2008): Sub-terminal sequences modulating IS30 transposition in vivo and in vitro. J.Mol.Biol.2008 Jan 11;375(2):337-52. Epub 2007 Oct 23.
Ferg M, Sanges R, Gehrig J, Kiss J,Bauer M, Lovas A, Szabo M,Yang L, Straehle U, Pankratz MJ, Olasz F, Stupka E, Müller F. (2007): The TATA-binding protein regulates maternal mRNA degradation and differential zygotic transcription in zebrafish. EMBO J. Sep 5;26(17):3945-56. Epub 2007 Aug 16;
János Kiss, Zita Nagy,Gábor Tóth, György Botond Kiss, Júlia Jakab, Michael Chandler, Ferenc Olasz:(2007): Transposition and target specificity of the typical IS30 family element IS1655 from Neisseria meningitidis.Molec. Microbiol. 63 (6): 1731-1747 MAR 2007
Nagy Zita,Mónika Szabó,Michael Chandler and Ferenc Olasz(2004): Analysis of the N-terminal DNA binding domain of the IS30 transposase. Molec. Microbiol. 54(2):478-88.
Kiss János, Mónika Szabó and Ferenc Olasz(2003): Site-specific recombination by the DDE-family member IS30 transposase. Proc Natl Acad Sci U S A.(100)25:1500-05.
Olasz Ferenc, Tamás Fischer, Mónika Szabó, Zita Nagy and János Kiss(2003).: Gene conversion in transposition of Escherichia coli elementIS30. J. Mol. Biol. 334/5 967-978.
Szabó Mónika,Ferenc Müller, János Kiss,Carolin Balduf, Uwe Strähle and Ferenc Olasz(2003): Trans-kingdom transposition and gene targeting mediated by the prokaryotic mobile element IS30FEBS Lett. 550(1-3):46-50.
Nagy, Z. Szeverényi, I. Farkas, T. Olasz, F. and Kiss J.(2003): Detection and analysis of transpositionally active head-to-tail dimers in three additional E. coliIS elements. Microbiology 149: 1297-1310
Kiss, J. and Olasz, F.(1999): Formation and transposition of the covalently closed IS30 circle: the relation between tandem dimers and monomeric circles. Molec. Microbiol. 34(1) 37-52
Olasz, F., Kiss, J.,König, P., Buzás, Zs., Stalder, R. and Arber W. (1998): Target specificity of insertion element IS30. Molec. Microbiol. 28(4) 691-704
Olasz, F., Farkas, T., Kiss, J.,Arini, A. and Arber W. (1997): Terminal inverted repeats of insertion sequence IS30 serve as target for transposition. J. Bacteriol. 179:7551-7558
Farkas, T, Kiss J, Olasz F(1996) The construction and characterization of an effective transpositional system based on IS30. FEBS Letters 390:53-58
Patent
Mónika Szabó, János Kiss, Ferenc Olasz, (ABC, Gödöllő, Hungary), Ferenc Müller, Lászlo Tora, Uwe Strähle (IGBMC, Strasbourg, France). European Patent application, No 01402754.4. Site-directed recombinase fusion proteins and corresponding polynucleotides, vectors and kits, and their uses for site-directed DNA recombination.